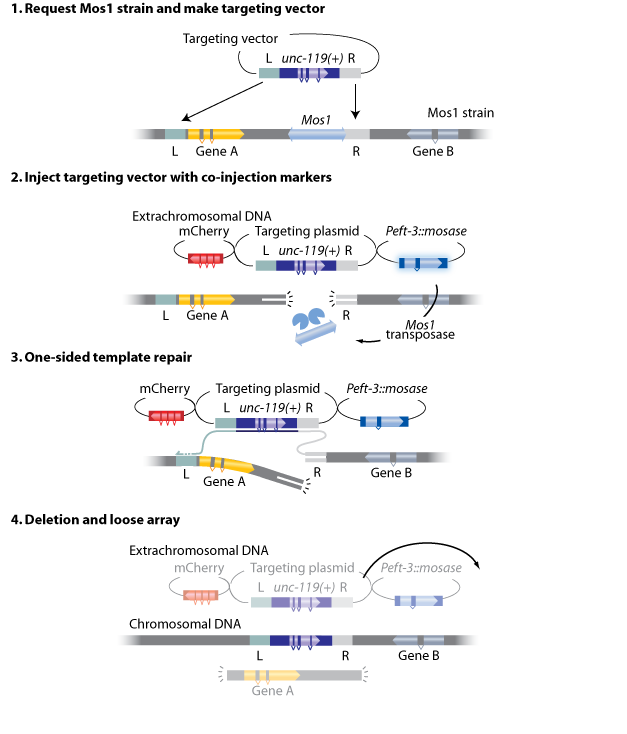
About Mos1-mediated Deletion (mosDEL)
mosDEL is a method to generate targeted deletions in the genome of C. elegans. The method has the advantages:
- Deletions are generated with selection markers
- The exact endpoints of a deletion can be specified
- Essential genes can be deleted and balanced at the same time
The method works by breaking a chromosome at a particular location by excising a Mos1 transposon. The excision creates a double-strand DNA break that is repaired by the cell. If there is a DNA template present with homology to one side of the breakpoint then the repair process will initiate repair from that side. If the repair template also contains homology to another nearby (within 25 kb) genomic region then the intervening genomic region is removed in the repair process. We have incorporated a positive selection marker to select for deletion events (typically unc-119 or NeoR) and fluorescent markers to select against extra-chromosomal arrays.
Requirements
- A Mos1 element within 25 kb of the gene/region to be deleted.
- Building a targeting vector with appropriate homology regions and selection markers.