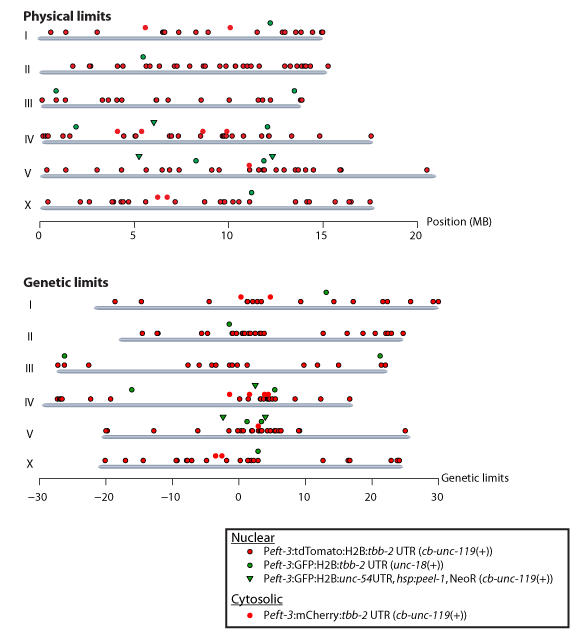
Overview of fluorescent marker strains (click here for printable pdf)
Use search box to limit strains to chromosomes (LG1, LG2 etc) or fluorophores (GFP, mCherry, tdTomato).
Please request strains from the CGC.
Strain column has link to the strain info at the CGC.
Please note that there are a couple of columns to the right of “insertion marker” in the table. Scroll to the right to see the exact genomic sequence that flanks the insertion site.
If you find the strains useful please consider citing “Frøkjær-Jensen C, Davis MW, Sarov M, Taylor J, Flibotte S, LaBella M, Pozniakovski A, Moerman DG and Jorgensen EM. Random and targeted transgene insertion in C. elegans using a modified Mos1 transposon. Nature Methods. (In press)”
Single marker strains
| Chr. | Genetic pos. | Position (WB230) | Promoter | Fluorophore | Allele | Strain | Insertion marker(s) | Genotype | Flanking sequence | Genomic environment | Comments |
|---|---|---|---|---|---|---|---|---|---|---|---|
| LG1 | I:-0.03 | 5,383,512 | Peft-3 | GFP(NLS) | oxTi995 | EG8934 | PuroR | oxTi995 I | (N)TGTGTCTGGAAAAATGATGTCATATCGCACT | In pqn-44 (one isoform) | Correction Feb 8, 2017: Genotype corrected to indicate insertion into N2 and NOT unc-119(ed3) |
| LG1 | I:-0.73 | 4,787,105 | Peft-3 | GFP(NLS) | oxTi951 | EG8899 | cbr-unc-119(+) | oxTi951 I ; unc-119(ed3) III | TATCTATCCGTTGCTTCTCCTTCCACCCAATACT | Intergenic | |
| LG1 | I:-1.28 | 4,470,840 | Peft-3 | GFP(NLS) | oxTi967 | EG8910 | cbr-unc-119(+) | oxTi967 I ; unc-119(ed3) III | TAAAAACTTTTTTAAACTAAATTTTGTTTAAAAAA | Intergenic | |
| LG1 | I:-1.50 | 4,339,885 | Peft-3 | GFP(NLS) | oxTi902 | EG8856 | cbr-unc-119(+) | oxTi902 I ; unc-119(ed3) III | TATTTAGTTGAGCTGAAGAAAATCGAAGATATG | In anc-1 | |
| LG1 | I:-14.58 | 1,572,773 | Peft-3 | GFP(NLS) | oxTi983 | EG8923 | HygroR | oxTi983 I | NATATATTTTCTCGAAATTTGGAACCGCCATAA | In src-1 | Correction Feb 8, 2017: Genotype corrected to indicate insertion into N2 and NOT unc-119(ed3) |
| LG1 | I:-14.67 | 1,455,659 | Peft-3 | tdTomato:H2B | oxTi638 | EG7832 | cbr-unc-119(+) | oxTi638 I ; unc-119(ed3) III | TACCTGCTAATTTTCAGTCAAGAACAAGAT | In spe-15. | |
| LG1 | I:-16.97 | 1,070,957 | Peft-3 | tdTomato:H2B | oxTi648 | EG7831 | cbr-unc-119(+) | oxTi648 I ; unc-119(ed3) III | TATATTTACGACCAGAACGTCTCATTTCGG | In gsa-1 | |
| LG1 | I:-18.09 | 866,406 | Peft-3 | GFP(NLS) | oxTi1016 | EG8952 | NeoR | oxTi1016 I | TATTATTTGAAATTATCGATTTTTTTTTCAAATTT | Y95B8A.8 | Correction Feb 8, 2017: Genotype corrected to indicate insertion into N2 and NOT unc-119(ed3) |
| LG1 | I:-18.63 | 657,302 | Peft-3 | tdTomato:H2B | oxTi550 | EG8004 | cbr-unc-119(+) | oxTi550 I ; unc-119(ed3) III | TAACCATTTCTCATTGTTTTTTTAAAATAC | In Y65B4A.1 | |
| LG1 | I:-18.96 | 525,573 | Peft-3 | GFP(NLS) | oxTi1009 | EG8947 | NeoR | oxTi1009 I | NATACAAGTTGCGCAAAGACCTTTCCAGTAG | Intergenic | Correction Feb 8, 2017: Genotype corrected to indicate insertion into N2 and NOT unc-119(ed3) |
| LG1 | I:-3.79 | 3,370,080 | Peft-3 | GFP(NLS) | oxTi934 | EG8886 | cbr-unc-119(+) | oxTi934 I ; unc-119(ed3) III | TNTTATTTGCCCCTTCTAGAATATTATAACTGTA | In W05F2.6 | |
| LG1 | I:-4.48 | 3,116,556 | Peft-3 | tdTomato:H2B | oxTi559 | EG7833 | cbr-unc-119(+) | oxTi559 I ; unc-119(ed3) III | TACTTCCGGATAGTTGGCGGTCGCATGTAG | In C45E1.4 | |
| LG1 | I:-7.69 | 2,698,143 | Peft-3 | GFP(NLS) | oxTi928 | EG8880 | cbr-unc-119(+) | oxTi928 I ; unc-119(ed3) III | TAAAAGTTAGTCTGCAAGAGTTCGCCGTGGC | In F32B5.2 | |
| LG1 | I:0.22 | 5,577,109 | Peft-3 | GFP(NLS) | oxTi895 | EG8850 | cbr-unc-119(+) | oxTi895 I ; unc-119(ed3) III | TATTTAGGTTACATCAGTATAAAAGGTTTGCAT | In klp-15 | |
| LG1 | I:1.23 | 6,585,618 | Peft-3 | tdTomato:H2B | oxTi556 | EG7835 | cbr-unc-119(+) | oxTi556 I ; unc-119(ed3) III | TATGCCGTTTTCCGATTGGTATTTCTAAAA | In try-6 | |
| LG1 | I:1.28 | 6,656,763 | Peft-3 | tdTomato:H2B | oxTi587 | EG7836 | cbr-unc-119(+) | oxTi587 I ; unc-119(ed3) III | TATGTATGTAATACCCGTTTTCCAAAAATA | In ZC581.2 | |
| LG1 | I:1.29 | 6,692,324 | Peft-3 | tdTomato:H2B | oxTi712 | EG7837 | cbr-unc-119(+) | oxTi712 I ; unc-119(ed3) III | TAAGAAAACTTCTATAAATCAATAAGCATA | Intergenic | |
| LG1 | I:1.58 | 6,966,807 | Peft-3 | GFP | umnTi4 | cbr-unc-119(+) | umnTi4 I; unc-119(ed3) III | Generated by the CGC | |||
| LG1 | I:1.60 | 6,999,163 | Peft-3 | GFP(NLS) | oxTi920 | EG8872 | cbr-unc-119(+) | oxTi920 I ; unc-119(ed3) III | TATATCTCAATCACTATAACTGAAAAATGCACACT | In C30F12.5 | |
| LG1 | I:1.67 | 7,049,861 | Peft-3 | GFP(NLS) | oxTi918 | EG8870 | cbr-unc-119(+) | oxTi918 I ; unc-119(ed3) III | TAATTATTTCATCATTCTTTAAAACTGCATCAT | Intergenic | |
| LG1 | I:13.92 | 12,687,214 | Peft-3 | GFP(NLS) | oxTi919 | EG8871 | cbr-unc-119(+) | oxTi919 I ; unc-119(ed3) III | TATACTCCGAAATACGCGGTTAATTTTCTCAAAA | In sra-25 | |
| LG1 | I:14.26 | 12,930,427 | Peft-3 | tdTomato:H2B | oxTi398 | EG7843 | cbr-unc-119(+) | oxTi398 I ; unc-119(ed3) III | TACTCTTCTCGAACTATGATGAAAAGTTGA | In Y18D10A.21 | |
| LG1 | I:17.17 | 13,063,874 | Peft-3 | tdTomato:H2B | oxTi413 | EG7844 | cbr-unc-119(+) | oxTi413 I ; unc-119(ed3) III | TATGTTTCGAGGTCACTAATAAGACTGTTT | Intergenic | |
| LG1 | I:19.25 | 13,351,226 | Peft-3 | GFP(NLS) | oxTi1002 | EG8941 | PuroR | oxTi1002 I | TATATGGCGTAACGGTCTCTCATCGCTCCGTAC | In Y40B1A.3 | Correction Feb 8, 2017: Genotype corrected to indicate insertion into N2 and NOT unc-119(ed3) |
| LG1 | I:2.07 | 7,436,394 | Peft-3 | tdTomato:H2B | oxTi718 | EG7838 | cbr-unc-119(+) | oxTi718 I ; unc-119(ed3) III | TATTATAATTGTAAAAAGAAGTAAATTTTT | In unc-13 | |
| LG1 | I:2.42 | 7,958,150 | Peft-3 | GFP(NLS) | oxTi977 | EG8919 | HygroR | oxTi977 I | TAGAAAAATCTGAATTCTTTCAGTTCTTCACAAT | In T22C1.8 | Correction Feb 8, 2017: Genotype corrected to indicate insertion into N2 and NOT unc-119(ed3) |
| LG1 | I:2.69 | 8,219,493 | Peft-3 | GFP(NLS) | oxTi946 | EG8896 | cbr-unc-119(+) | oxTi946 I ; unc-119(ed3) III | TAAGTGTATTTTATCGAGAAATTTTTAAAAAACG | Intergenic | |
| LG1 | I:2.75 | 8,346,811 | Peft-3 | tdTomato:H2B | oxTi623 | EG7839 | cbr-unc-119(+) | oxTi623 I ; unc-119(ed3) III | TATTCGGTGAAGAACAATCATGAGAAAAGA | Intergenic | |
| LG1 | I:21.62 | 13,636,871 | Peft-3 | tdTomato:H2B | oxTi571 | EG7845 | cbr-unc-119(+) | oxTi571 I ; unc-119(ed3) III | TATAAGGAAGGTTGCGAACTGAAATTTTTT | Intergenic | |
| LG1 | I:21.64 | 13,650,194 | Peft-3 | GFP(NLS) | oxTi1018 | EG8954 | NeoR | oxTi1018 I | TAGCTCTTATTATTATCTATATTTTGATAATTTACT | Intergenic | Correction Feb 8, 2017: Genotype corrected to indicate insertion into N2 and NOT unc-119(ed3) |
| LG1 | I:22.30 | 13,917,472 | Peft-3 | tdTomato:H2B | oxTi700 | EG7846 | cbr-unc-119(+) | oxTi700 I ; unc-119(ed3) III | TACAATCTACACTTTTCACAGTTTCCATAC | In clec-111 | |
| LG1 | I:22.34 | 13,940,945 | Peft-3 | GFP(NLS) | oxTi1022 | EG8958 | NeoR | oxTi1022 I | TAGTTGCATGCTACATTGATGACTTCAAAATTAA | In Y71A12B.8 | Correction Feb 8, 2017: Genotype corrected to indicate insertion into N2 and NOT unc-119(ed3) |
| LG1 | I:24.67 | 14,380,521 | Peft-3 | GFP(NLS) | oxTi926 | EG8878 | cbr-unc-119(+) | oxTi926 I ; unc-119(ed3) III | TACATATTGTACCACTGAACGTGTTCCTAAACTG | Intergenic | |
| LG1 | I:24.77 | 14,387,440 | Peft-3 | GFP(NLS) | oxTi981 | EG8922 | HygroR | oxTi981 I | TAAATTCCCGCATTTTTCGAAGATAAAAGCGAA | Intergenic | Correction Feb 8, 2017: Genotype corrected to indicate insertion into N2 and NOT unc-119(ed3) |
| LG1 | I:25.79 | 14,524,049 | Peft-3 | tdTomato:H2B | oxTi723 | EG7847 | cbr-unc-119(+) | oxTi723 I ; unc-119(ed3) III | TAAGTTCTGCAAAACTAGTTTCCCATTGAA | Intergenic | |
| LG1 | I:25.92 | 14,645,019 | Peft-3 | GFP(NLS) | oxTi993 | EG8932 | PuroR | oxTi993 I | TATTTTAGAAGTTTATCTAAATTCAGTTCGTTGA | In Y105E8B.7 | Correction Feb 8, 2017: Genotype corrected to indicate insertion into N2 and NOT unc-119(ed3) |
| LG1 | I:29.18 | 15,003,086 | Peft-3 | tdTomato:H2B | oxTi626 | EG7848 | cbr-unc-119(+) | oxTi626 I ; unc-119(ed3) III | TAATTGGTGTGACGTCACATCAGTCAATAG | In mtd-1 | |
| LG1 | I:29.97 | 15,070,626 | Peft-3 | tdTomato:H2B | oxTi730 | EG7854 | cbr-unc-119(+) | oxTi730 I ; unc-119(ed3) III | TAGCCGCACGAGATTGAGCGATAACAGGTC | In 18S rRNA. Non-unique. Maps to LG1: 15,063,429 and 15,070,626 | |
| LG1 | I:3.35 | 8,993,439 | Peft-3 | tdTomato:H2B | oxTi590 | EG7840 | cbr-unc-119(+) | oxTi590 I ; unc-119(ed3) III | TAAATAATTTCCAGCTGCAGCTAAACTTCG | In F32H2.8 | |
| LG1 | I:3.44 | 9,050,831 | Peft-3 | GFP(NLS) | oxTi923 | EG8875 | cbr-unc-119(+) | oxTi923 I ; unc-119(ed3) III | TACCAATCAAGAAATCTCCATCAGAATTATACA | In hgo-1 | |
| LG1 | I:3.91 | 9,803,196 | Peft-3 | GFP(NLS) | oxTi959 | EG8904 | cbr-unc-119(+) | oxTi959 I ; unc-119(ed3) III | TAAACCCAACTTTTGCAATGGAATGGCACCGAA | Intergenic | |
| LG1 | I:30.0 | 15,065,881 | Peft-3 | GFP | umnTi3 | cbr-unc-119(+) | umnTi3 I; unc-119(ed3) III | Generated by the CGC | |||
| LG1 | I:4.72 | 10,166,146 | Peft-3 | mCherry | oxTi302 | EG7841 | cbr-unc-119(+) | oxTi302 I ; unc-119(ed3) III | TATATCGAGAGCCAACGGGAAACAATTGGG | Intergenic | |
| LG1 | I:5.06 | 10,713,324 | Peft-3 | GFP(NLS) | oxTi979 | EG8920 | HygroR | oxTi979 I | NAAATTCCCCCGTATAAAAAGAAAGCGGCAACC | In B0205.1 | Correction Feb 8, 2017: Genotype corrected to indicate insertion into N2 and NOT unc-119(ed3) |
| LG1 | I:7.93 | 11,259,211 | Peft-3 | GFP(NLS) | oxTi925 | EG8877 | cbr-unc-119(+) | oxTi925 I ; unc-119(ed3) III | NACATCATCTCAAGGGAGATTGCCAAAACAAT | In F36D1.8 (promoter) | |
| LG1 | I:8.72 | 11,460,740 | Peft-3 | GFP(NLS) | oxTi917 | EG8869 | cbr-unc-119(+) | oxTi917 I ; unc-119(ed3) III | TATAATTTCCGGCATGGTCGGAATAACGCCG | Intergenic | |
| LG1 | I:9.28 | 11,592,512 | Peft-3 | tdTomato:H2B + gaIs283 | oxTi331 | EG7213 | cbr-unc-119(+) | oxTi331 I ; gaIs283 | TACAGTACCCCTGCTTTGCGGCGGGACCTA | In unc-75 | |
| LG1 | I:9.62 | 11,788,128 | Peft-3 | tdTomato:H2B | oxTi653 | EG7842 | cbr-unc-119(+) | oxTi653 I ; unc-119(ed3) III | TAGGCATGGGCTCAGGCTTCGACTTAGGCT | In vab-10 | |
| LG2 | II:-0.38 | 6,311,694 | Peft-3 | tdTomato:H2B | oxTi564 | EG7866 | cbr-unc-119(+) | oxTi564 II ; unc-119(ed3) III | TATTTTACATTTACTTTATCTGATGACAAC | Intergenic | |
| LG2 | II:-0.61 | 6,035,748 | Peft-3 | GFP(NLS) | oxTi891 | EG8846 | cbr-unc-119(+) | oxTi891 II ; unc-119(ed3) III | TACATACATGTAAAATAATTAAGCTAGACTTCAA | Intergenic | |
| LG2 | II:-0.92 | 5,811,246 | Peft-3 | tdTomato:H2B | oxTi617 | EG7865 | cbr-unc-119(+) | oxTi617 II ; unc-119(ed3) III | TACTTCAAGATATCAGTATCAGGTACATCG | In ZK1248.5 | |
| LG2 | II:-1.02 | 5,612,590 | Peft-3 | tdTomato:H2B | oxTi726 | EG7864 | cbr-unc-119(+) | oxTi726 II ; unc-119(ed3) III | TACATACATACCTATAATATATTCAGAACA | Intergenic | |
| LG2 | II:-1.99 | 5,151,443 | Peft-3 | GFP(NLS) | oxTi1011 | EG8949 | NeoR | oxTi1011 II | TACACTAAAATTATGGTAGTTTTTCCTCTTCTTT | Intergenic | Correction Feb 8, 2017: Genotype corrected to indicate insertion into N2 and NOT unc-119(ed3) |
| LG2 | II:-11.63 | 2,770,138 | Peft-3 | GFP(NLS) | oxTi974 | EG8916 | HygroR | oxTi974 II | TAACGTGAGAGCTTCTCTAGTATCATCTTTGGA | In fbxb-105 | Correction Feb 8, 2017: Genotype corrected to indicate insertion into N2 and NOT unc-119(ed3) |
| LG2 | II:-12.17 | 2,671,028 | Peft-3 | tdTomato:H2B | oxTi677 | EG7860 | cbr-unc-119(+) | oxTi677 II ; unc-119(ed3) III | TATAACATTTTTCGAATGTTAAGTTTCATG | In sri-57 | |
| LG2 | II:-12.31 | 2,609,730 | Peft-3 | tdTomato:H2B | oxTi402 | EG7859 | cbr-unc-119(+) | oxTi402 II ; unc-119(ed3) III | TAAATAAAAAAGTTTCAAAAATTTTTTATA | Intergenic | |
| LG2 | II:-14.32 | 1,834,746 | Pmyo-3 | GFP(NLS) | oxTi886 | EG8841 | cbr-unc-119(+) | oxTi886 II ; unc-119(ed3) III | TAACATTGCAGACTTCACTTGGGT | In T08E11.8 | |
| LG2 | II:-14.52 | 1,725,214 | Peft-3 | tdTomato:H2B | oxTi729 | EG7858 | cbr-unc-119(+) | oxTi729 II ; unc-119(ed3) III | TAGATACGATTTGTTTTAGCTTCAGTCTCA | In fbxb-11. | |
| LG2 | II:-15.12 | 1,639,547 | Peft-3 | GFP(NLS) | oxTi984 | EG8924 | HygroR | oxTi984 II | TATATACAGCATGAAGTGAAGAATATAATTTGCAG | In srh-72 | Correction Feb 8, 2017: Genotype corrected to indicate insertion into N2 and NOT unc-119(ed3) |
| LG2 | II:-15.44 | 1,470,341 | Peft-3 | tdTomato:H2B | oxTi624 | EG7856 | cbr-unc-119(+) | oxTi624 II ; unc-119(ed3) III | TAGACTAGTACTTACCACTATACGATTTGA | In col-70 | |
| LG2 | II:-15.96 | 136,399 | Peft-3 | GFP(NLS) | oxTi992 | EG8931 | HygroR | oxTi992 II | TATTAATCGATGAGCACAATTGGGTAGTAGGA | In K10B4.1 | Correction Feb 8, 2017: Genotype corrected to indicate insertion into N2 and NOT unc-119(ed3) |
| LG2 | II:-16.19 | 93,136 | Peft-3 | tdTomato:H2B | oxTi724 | EG7855 | cbr-unc-119(+) | oxTi724 II ; unc-119(ed3) III | TAGACCAATCTGTTCCAGAAGATACCATAA | In C50D2.7 | |
| LG2 | II:-17.95 | 49,663 | Peft-3 | GFP(NLS) | oxTi944 | EG8894 | cbr-unc-119(+) | oxTi944 II ; unc-119(ed3) III | TAGCATCTTCGTCGCTTTCAATTTCAGGTATT | In C23H3.5 | |
| LG2 | II:-2.02 | 5,130,813 | Peft-3 | GFP | oxTi877 | EG8834 | cbr-unc-119(+) | oxTi877 II ; unc-119(ed3) III | TATTGTATTCAAAATTTGCAAGCTACATAGCAG | In ggr-3 | |
| LG2 | II:-2.60 | 5,011,388 | Peft-3 | GFP(NLS) | oxTi941 | EG8892 | cbr-unc-119(+) | oxTi941 II ; unc-119(ed3) III | TAAACATTTTTCAGAATATTTTGAACAACAAAAG | Intergenic | |
| LG2 | II:-4.73 | 4,382,212 | Peft-3 | tdTomato:H2B | oxTi717 | EG7863 | cbr-unc-119(+) | oxTi717 II ; unc-119(ed3) III | TACATCATCTTTCCCATATCACATTTTCGT | In fkh-6 | |
| LG2 | II:-5.62 | 4,091,582 | Peft-3 | tdTomato:H2B | oxTi647 | EG7862 | cbr-unc-119(+) | oxTi647 II ; unc-119(ed3) III | TACGTGCCTGCCTACCGCCTGCATTTTGGC | Intergenic | |
| LG2 | II:0.12 | 6,816,385 | Peft-3 | GFP(NLS) | oxTi939 | EG8890 | cbr-unc-119(+) | oxTi939 II ; unc-119(ed3) III | TAACTTCTAAAACTGATTTTATAAAAAATATTT | Intergenic (snt-1 UTR) | |
| LG2 | II:0.13 | 6,865,705 | Pvha-6 | GFP(NLS) | oxTi882 | EG8838 | cbr-unc-119(+) | oxTi882 II ; unc-119(ed3) III | TAATTCCCGCATAGTGCGCGGCACTCAGCGG | In ncRNA C44B7.21 | |
| LG2 | II:0.49 | 7,110,236 | Peft-3 | tdTomato:H2B | oxTi394 | EG7867 | cbr-unc-119(+) | oxTi394 II ; unc-119(ed3) III | TATAACGCTCACCGTTGATGAACATCCATT | Intergenic | |
| LG2 | II:0.50 | 7,247,795 | Peft-3 | tdTomato:H2B | oxTi629 | EG7868 | cbr-unc-119(+) | oxTi629 II ; unc-119(ed3) III | TATTAACACTGGTTCAAATGCGCGATGCAG | In F32A5.2 | |
| LG2 | II:0.50 | 7,169,878 | Peft-3 | GFP(NLS) | oxTi904 | EG8858 | cbr-unc-119(+) | oxTi904 II ; unc-119(ed3) III | TATATCAATTGTTATTAGTCATTAAAAAAAACT | In dyf-13 | |
| LG2 | II:0.56 | 7,767,187 | Peft-3 | GFP(NLS) | oxTi940 | EG8891 | cbr-unc-119(+) | oxTi940 II ; unc-119(ed3) III | TACATCCTAATATTAAAAAAAACCACTCTGAAA | In C06A8.8 | |
| LG2 | II:0.61 | 7,918,066 | Peft-3 | tdTomato:H2B | oxTi582 | EG7869 | cbr-unc-119(+) | oxTi582 II ; unc-119(ed3) III | TAGAATTCAGAAATAACTAGCAGACATGAA | In tag-341 | |
| LG2 | II:0.83 | 8,641,549 | Peft-3 | tdTomato:H2B | oxTi540 | EG7870 | cbr-unc-119(+) | oxTi540 II ; unc-119(ed3) III | TATAAGTAGTTTTTAGTAGTTACTGTCTCA | In D2085.7 | |
| LG2 | II:0.84 | 8,679,134 | Peft-3 | GFP(NLS) | oxTi976 | EG8918 | HygroR | oxTi976 II | NACTTGATATTAACAGAAGCTGTTAAACAAT | In D2085.5 | Correction Feb 8, 2017: Genotype corrected to indicate insertion into N2 and NOT unc-119(ed3) |
| LG2 | II:0.87 | 8,739,383 | Peft-3 | tdTomato:H2B | oxTi548 | EG7871 | cbr-unc-119(+) | oxTi548 II ; unc-119(ed3) III | TATTTCAAAGACATTCCTGTTCGCGTTCCC | Intergenic | |
| LG2 | II:0.87 | 8,723,252 | Peft-3 | GFP(NLS) | oxTi990 | EG8929 | HygroR | oxTi990 II | TACTTCCCATCAGCTTAGTAACGTCTTGGAC | In trcs-2 | Correction Feb 8, 2017: Genotype corrected to indicate insertion into N2 and NOT unc-119(ed3) |
| LG2 | II:1.48 | 9,513,818 | Peft-3 | tdTomato:H2B | oxTi573 | EG7872 | cbr-unc-119(+) | oxTi573 II ; unc-119(ed3) III | TATATTCATTTGCTATAATAGCATCAGAGA | In gcy-1 | |
| LG2 | II:1.73 | 9,839,241 | Peft-3 | tdTomato:H2B | oxTi609 | EG7873 | cbr-unc-119(+) | oxTi609 II ; unc-119(ed3) III | TATTAGTGAATCAAGTTCGAAAAATCTATT | Intergenic | |
| LG2 | II:10.05 | 12,462,544 | Peft-3 | GFP(NLS) | oxTi930 | EG8882 | cbr-unc-119(+) | oxTi930 II ; unc-119(ed3) III | TATTTCTGAAGTACTGAATCACCTTAAAAA | In F15A4.2 | |
| LG2 | II:10.79 | 12,708,603 | Peft-3 | GFP(NLS) | oxTi1010 | EG8948 | NeoR | oxTi1010 II | TAATATGTACACTAGAAAATTGTTGGAAGA | In tbc-17 | Correction Feb 8, 2017: Genotype corrected to indicate insertion into N2 and NOT unc-119(ed3) |
| LG2 | II:12.65 | 12,954,556 | Peft-3 | tdTomato:H2B | oxTi296 | EG7879 | cbr-unc-119(+) | oxTi296 II ; unc-119(ed3) III | TAATACGGGGGGAGATTTTCAGCATTTCAG | In Y38F1A | |
| LG2 | II:16.25 | 13,245,885 | Peft-3 | tdTomato:H2B | oxTi589 | EG7880 | cbr-unc-119(+) | oxTi589 II ; unc-119(ed3) III | TATTTTCACAGGAAATTCCTTTCGTGGGTA | In F15D4.5 | |
| LG2 | II:18.17 | 13,576,058 | Peft-3 | GFP(NLS) | oxTi969 | EG8912 | cbr-unc-119(+) | oxTi969 II ; unc-119(ed3) III | TATTGCCGGAAATTCGACTGAAAACCGGCAAT | In lin-38 | |
| LG2 | II:18.64 | 13,601,638 | Peft-3 | tdTomato:H2B | oxTi409 | EG7881 | cbr-unc-119(+) | oxTi409 II ; unc-119(ed3) III ; him-5(e1490) V | TATTAAGATTATTTCGGCGAGTTTCACATA | In csp-1 | |
| LG2 | II:2.43 | 10,335,848 | Peft-3 | tdTomato:H2B | oxTi310 | EG7828 | cbr-unc-119(+) | oxTi310 II ; unc-119(ed3) III | TAAGTGTATTTTTGCTATTTTTTTAATCATAA | Intergenic | |
| LG2 | II:2.45 | 10,347,029 | Peft-3 | tdTomato:H2B | oxTi714 | EG7874 | cbr-unc-119(+) | oxTi714 II ; unc-119(ed3) III | TAAAATAACTTTTATCGTAAGTTTAAACAA | Intergenic | |
| LG2 | II:2.59 | 10,460,500 | Pmyo-3 | GFP(NLS) | oxTi887 | EG8842 | cbr-unc-119(+) | oxTi887 II ; unc-119(ed3) III | TAAGTGNTTCCGGTAAAGTGTTGAAAAAGT | In ZK673.11 | |
| LG2 | II:20.30 | 13,818,654 | Peft-3 | GFP(NLS) | oxTi943 | EG8893 | cbr-unc-119(+) | oxTi943 II ; unc-119(ed3) III | TACTTGGTAACGGCCTTGGTTCCCTCAGACA | in histone (not unique) | |
| LG2 | II:20.47 | 13,851,592 | Peft-3 | tdTomato:H2B | oxTi657 | EG7882 | cbr-unc-119(+) | oxTi657 II ; unc-119(ed3) III | TAAAATTTAACATATTTAAAAAAAAATTTT | Intergenic | |
| LG2 | II:22.40 | 14,074,299 | Peft-3 | tdTomato:H2B | oxTi607 | EG7884 | cbr-unc-119(+) | oxTi607 II ; unc-119(ed3) III | TATGCCCCCCAAGATACACCCCAGTTTGTC | In xrn-1 | |
| LG2 | II:22.90 | 14,310,914 | Peft-3 | tdTomato:H2B | oxTi721 | EG7885 | cbr-unc-119(+) | oxTi721 II ; unc-119(ed3) III | TAAAATCGATTAATTTTTTTTCCCGGAAAA | Intergenic | |
| LG2 | II:22.90 | 14,289,311 | Peft-3 | GFP(NLS) | oxTi924 | EG8876 | cbr-unc-119(+) | oxTi924 II ; unc-119(ed3) III | (N)ACTACTTCCCCTACGCCCAGTAAAGT | In srh-42 | |
| LG2 | II:24.67 | 15,249,353 | Peft-3 | [tdTomato:H2B | oxTi399 | EG7886 | cbr-unc-119(+) | oxTi399 II ; unc-119(ed3) III | TATTATAATTAATTAGAAATTTCTAGAAAT | In tig-3 | |
| LG2 | II:3.08 | 10,546,809 | Peft-3 | GFP(NLS) | oxTi957 | EG8903 | cbr-unc-119(+) | oxTi957 II ; unc-119(ed3) III | TACTGTGTAGTAGTAAATCAATCAAATGTCT | in mnk-1 | |
| LG2 | II:3.13 | 10,754,369 | Peft-3 | tdTomato:H2B | oxTi574 | EG7875 | cbr-unc-119(+) | oxTi574 II ; unc-119(ed3) III | TAAGTTTTTTTGAACATTTTATTGGTAGCT | In mpz-1 | |
| LG2 | II:3.13 | 10,966,694 | Peft-3 | tdTomato:H2B | oxTi722 | EG7876 | cbr-unc-119(+) | oxTi722 II ; unc-119(ed3) III | TATGTGCAAATTTTGAATTTTTGACTGAAA | Intergenic | |
| LG2 | II:3.15 | 11,029,447 | Peft-3 | GFP(NLS) | oxTi903 | EG8857 | cbr-unc-119(+) | oxTi903 II ; unc-119(ed3) III | TATCTTGAAGGGTAATATGAACATTTACGAGCTT | In nlp-18 | |
| LG2 | II:3.16 | 11,056,681 | Peft-3 | GFP(NLS) | oxTi929 | EG8881 | cbr-unc-119(+) | oxTi929 II ; unc-119(ed3) III | TAAGTACTTTTGTTGTCATTTTGTTTTGGACG | In F33A8.6 | |
| LG2 | II:3.34 | 11,168,622 | Peft-3 | tdTomato:H2B | oxTi583 | EG7877 | cbr-unc-119(+) | oxTi583 II ; unc-119(ed3) III | TAATATTTCAAGGAAGGGCCGGCCTGCTAG | Intergenic | |
| LG2 | II:3.49 | 11,469,692 | Peft-3 | GFP(NLS) | oxTi931 | EG8883 | cbr-unc-119(+) | oxTi931 II ; unc-119(ed3) III | TATTTTACAATTAGAATTTTTCGCTCATTTTCCGC | Intergenic | |
| LG2 | II:3.79 | 11,601,311 | Peft-3 | tdTomato:H2B | oxTi720 | EG7878 | cbr-unc-119(+) | oxTi720 II ; unc-119(ed3) III | TAGTATGCCAATTAAAAATCAATTAGTTTT | Intergenic | |
| LG3 | II:-7.66 | 3,207,601 | Peft-3 | tdTomato:H2B | oxTi706 | EG7890 | cbr-unc-119(+) | unc-119(ed3) oxTi706 III | TATTAGGTGACGATC | Intergenic | |
| LG3 | III:-0.26 | 8,440,662 | Peft-3 | tdTomato:H2B | oxTi719 | EG7897 | cbr-unc-119(+) | oxTi719 unc-119(ed3) III | TAAGTTTCAATAAGCAATAGCAATAAGTTT | In ZK1236.7 | |
| LG3 | III:-0.35 | 8,258,097 | Peft-3 | GFP(NLS) | oxTi962 | EG8906 | cbr-unc-119(+) | oxTi962 unc-119(ed3) III | TAACTACTTTTACGAACTGTACATTATAGCAGAA | Intergenic | |
| LG3 | III:-0.36 | 8,212,287 | Peft-3 | GFP(NLS) | oxTi897 | EG8851 | cbr-unc-119(+) | oxTi897 unc-119(ed3) III | TAATATCGTGTAAGTACTGTGACAACAAAGAAG | Intergenic | |
| LG3 | III:-0.45 | 8,004,972 | Peft-3 | GFP(NLS) | oxTi1001 | EG8940 | PuroR | oxTi1001 | TACTCCTTCTTCATNACTTAACAATCAACCCATN | In C06G4.6 | Correction Feb 8, 2017: Genotype corrected to indicate insertion into N2 and NOT unc-119(ed3) |
| LG3 | III:-0.93 | 6,852,302 | Peft-3 | GFP(NLS) | oxTi892 | EG8847 | cbr-unc-119(+) | oxTi892 unc-119(ed3) III | TATATTTTATGTTCTTGATC | In srv-1 | |
| LG3 | III:-1.01 | 6,689,580 | Peft-3 | tdTomato:H2B | oxTi627 | EG7896 | cbr-unc-119(+) | oxTi627 unc-119(ed3) III | TATTACTTGATTGCTCATCTCAAAGCTTAA | In rbf-1 | |
| LG3 | III:-1.41 | 6,196,016 | Peft-3 | GFP | oxTi872 | EG8829 | cbr-unc-119(+) | unc-119(ed3) oxTi872 III | TATAAACTTTGAAAAAAAAACCGGCAAACTTTT | In C23G10.7 3'UTR | |
| LG3 | III:-1.42 | 6,076,840 | Peft-3 | tdTomato:H2B | oxTi411 | EG7895 | cbr-unc-119(+) | oxTi411 unc-119(ed3) III | TAAATGTAATTTTTTTTTTTAACTAAACCG | Intergenic | |
| LG3 | III:-1.42 | 6,058,432 | Peft-3 | tdTomato:H2B | oxTi603 | EG7894 | cbr-unc-119(+) | oxTi603 unc-119(ed3) III | TACACCTCATTTTCCTGTTCAAAATCTAGA | Intergenic | |
| LG3 | III:-1.43 | 5,935,821 | Peft-3 | GFP | umnTi6 | cbr-unc-119(+) | umnTi6 unc-119(ed3) III | Generated by the CGC | |||
| LG3 | III:-14.69 | 2,365,382 | Peft-3 | GFP(NLS) | oxTi1005 | EG8943 | PuroR | oxTi1005 | TAAAATGAAAAAGTAACGCTTTTAAACGATTTCG | In srd-69 | Correction Feb 8, 2017: Genotype corrected to indicate insertion into N2 and NOT unc-119(ed3) |
| LG3 | III:-15.41 | 2,077,990 | Peft-3 | GFP(NLS) | oxTi945 | EG8895 | cbr-unc-119(+) | oxTi945 unc-119(ed3) III | TACCTGTTGAATTGGAATTCAAGACAAAAACC | Intergenic | |
| LG3 | III:-22.58 | 1,247,913 | Peft-3 | tdTomato:H2B + gaIs283 | oxTi330 | EG7212 | cbr-unc-119(+) | oxTi330 III ; gaIs283 | TAAAACCACGGAATATGGAAAAAGTTCCCG | In fbxa-76 | |
| LG3 | III:-26.23 | 763,007 | Peft-3 | tdTomato:H2B | oxTi641 | EG7888 | cbr-unc-119(+) | oxTi641 unc-119(ed3) III | TATGTGAATTTAGGTTTTATGTAAGACCAA | Intergenic | |
| LG3 | III:-27.22 | 15,501 | Peft-3 | tdTomato:H2B | oxTi731 | EG7887 | cbr-unc-119(+) | oxTi731 unc-119(ed3) III | TAGAAACGTGTAGCCTTATCAATGTATCAC | Intergenic | |
| LG3 | III:-3.17 | 4,454,757 | Peft-3 | GFP(NLS) | oxTi935 | EG8887 | cbr-unc-119(+) | oxTi935 unc-119(ed3) III | NAGAAGATATGTATAATGCTAAAATTCTTTTTTT | Intergenic | |
| LG3 | III:-3.47 | 4,238,204 | Peft-3 | tdTomato:H2B | oxTi615 | EG7893 | cbr-unc-119(+) | oxTi615 unc-119(ed3) III | TAAACAATTTTTTTAGTGGGCGCTCAATAG | Intergenic | |
| LG3 | III:-3.80 | 4,102,940 | Peft-3 | GFP(NLS) | oxTi1024 | EG8960 | NeoR | oxTi1024 | TAGGTAAAAATTCCATCAGTTTGAATTGTGTCT | Intergenic | Correction Feb 8, 2017: Genotype corrected to indicate insertion into N2 and NOT unc-119(ed3) |
| LG3 | III:-3.94 | 4,060,854 | Peft-3 | GFP(NLS) | oxTi975 | EG8917 | HygroR | oxTi975 | TACATGGTTGAACCATTAACTGAGTNGGTTAA | Intergenic/pseudogene | Correction Feb 8, 2017: Genotype corrected to indicate insertion into N2 and NOT unc-119(ed3) |
| LG3 | III:-4.08 | 3,974,175 | Peft-3 | tdTomato:H2B | oxTi612 | EG7892 | cbr-unc-119(+) | oxTi612 unc-119(ed3) III | TATGTCAGTGAACACATAACCAAACTGGTA | Intergenic | |
| LG3 | III:-4.25 | 3,771,500 | Peft-3 | GFP | umnTi1 | cbr-unc-119(+) | umnTi1 unc-119(ed3) III | Generated by the CGC | |||
| LG3 | III:-5.98 | 3,524,591 | Peft-3 | tdTomato:H2B | oxTi544 | EG7891 | cbr-unc-119(+) | oxTi544 unc-119(ed3) III | TAACATCATTTTGCAGGGCTATTATTAATG | In pitp-1 | |
| LG3 | III:0.03 | 8,915,141 | Peft-3 | GFP(NLS) | oxTi956 | EG8902 | cbr-unc-119(+) | oxTi956 III unc-119(ed3) III | TACACCATTACAGGAAGCTCTATCAAATTCATC | In trxr-2 | |
| LG3 | III:1.23 | 9,927,302 | Peft-3 | tdTomato:H2B | oxTi619 | EG7898 | cbr-unc-119(+) | oxTi619 unc-119(ed3) III | TAAAATCAACGGCAGTCTAGACTGGAGA | Intergenic | |
| LG3 | III:11.80 | 11,696,626 | Peft-3 | tdTomato:H2B | oxTi546 | EG7900 | cbr-unc-119(+) | unc-119(ed3) oxTi546 III | TATTCACGTGGTGCCAGGCTGTCCCATTAC | In Y41C4A.6 | |
| LG3 | III:13.25 | 11,816,400 | Peft-3 | GFP | umnTi2 | cbr-unc-119(+) | umnTi2 unc-119(ed3) III | Generated by the CGC | |||
| LG3 | III:14.97 | 12,113,652 | Peft-3 | tdTomato:H2B | oxTi539 | EG7901 | cbr-unc-119(+) | unc-119(ed3) oxTi539 III | TATATATTACTGATTTAGAATGCTCTTGCTC | Intergenic | |
| LG3 | III:19.10 | 12,863,287 | Peft-3 | GFP(NLS) | oxTi989 | EG8928 | HygroR | oxTi989 | TATTTCTTCCCCTTTTTATTTGCTTTTTTGCATCA | Intergenic | Correction Feb 8, 2017: Genotype corrected to indicate insertion into N2 and NOT unc-119(ed3) |
| LG3 | III:21.39 | 13,706,630 | Peft-3 | tdTomato:H2B | oxTi713 | EG7902 | cbr-unc-119(+) | unc-119(ed3) oxTi713 III | TAGATAGAGTGAAGTAGGTTAAAAAAGAGC | In lit-1 | |
| LG3 | III:22.01 | 13,780,498 | Peft-3 | tdTomato:H2B | oxTi715 | EG7903 | cbr-unc-119(+) | unc-119(ed3) oxTi715 III | TAGAAGATGAATTAAAAGTGTTACATTTAG | In pot-3 | |
| LG4 | IV:-1.41 | 3,951,234 | Peft-3 | mCherry | oxTi420 | EG7910 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi420 IV | TATGATCTTTCGTCACGAACAAATGGACTA | Intergenic | |
| LG4 | IV:-16.25 | 1,691,577 | Peft-3 | GFP(NLS) | oxTi927 | EG8879 | cbr-unc-119(+) | unc-119(ed3) III ;oxTi927 IV | TAATTGATTGCGTCTAACTTAATTCAATATAGCT | Intergenic | |
| LG4 | IV:-19.29 | 1,425,441 | Peft-3 | tdTomato:H2B | oxTi670 | EG7909 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi670 IV | TATACATCAGATTAGAGCAAAATCAATCAT | Intergenic | |
| LG4 | IV:-22.29 | 1,064,173 | Peft-3 | tdTomato:H2B | oxTi636 | EG7908 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi636 IV | TAAATTTCTAAGTATCAAATCATAGCCAAG | Intergenic | |
| LG4 | IV:-25.76 | 563,308 | Peft-3 | GFP(NLS) | oxTi914 | EG8867 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi914 IV | TATGCCAGCAGACGTTCTGTGTTAATACAAACA | F56A11.7 | |
| LG4 | IV:-26.58 | 278,064 | Peft-3 | tdTomato:H2B | oxTi408 | EG7907 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi408 IV ; him-5(e1490) V | TACACTTGTCATGCAACTAATGTTGCCGGG | In sma-10 | |
| LG4 | IV:-26.79 | 239,554 | Peft-3 | tdTomato:H2B | oxTi608 | EG7906 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi608 IV | TAGATGTTTCTCCACTAAAAACGCTAATCG | In fnta-1 | |
| LG4 | IV:-26.93 | 132,649 | Peft-3 | tdTomato:H2B | oxTi390 | EG7905 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi390 IV | TACATAAATTTTTAGTTATAATCACAAATT | In daf-1 | |
| LG4 | IV:-27.28 | 7,631 | Peft-3 | tdTomato:H2B | oxTi716 | EG7904 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi716 IV | TAGAGTACCCTAATGTATCCTTAAATTACC | In frm-5.2 or frm-5.1. | Non-unique. 7,631 or 40,248 Chr. IV. |
| LG4 | IV:-5.63 | 3,208,490 | Peft-3 | GFP(NLS) | oxTi915 | EG8868 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi915 IV | TAAACTCTTCAGCCTCACTCAAATTCTTCAACC | In puf-4 (pseudogene) | |
| LG4 | IV:0.09 | 4,281,636 | Peft-3 | tdTomato:H2B | oxTi705 | EG7911 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi705 IV | TATAGTACGACTGTATTAAAAGCAATCCAA | In F15E6.6 | |
| LG4 | IV:1.39 | 4,886,853 | Peft-3 | tdTomato:H2B | oxTi397 | EG7912 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi397 IV | TATACAGTTTCTGAATCTCTTTTGATTGTC | In str-166 | |
| LG4 | IV:1.39 | 4,909,581 | Peft-3 | tdTomato:H2B | oxTi563 | EG7913 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi563 IV | TATGTGTATACGTTTGAAGCTAGTAGTGTA | In str-159 | |
| LG4 | IV:1.39 | 4,922,692 | Peft-3 | tdTomato:H2B | oxTi622 | EG7914 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi622 IV | TAGTACTGTAGAGGTACTGTAGGAGTACTG | In Y9C9A.8 | Non-unique. 4,922,692 or 4,924,961 or 4,927,245 Chr. IV. |
| LG4 | IV:1.58 | 5,230,794 | Peft-3 | mCherry | oxTi419 | EG7915 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi419 IV | TATACTGTTCCGCTGGTAATGAAAACAATG | Intergenic | |
| LG4 | IV:1.82 | 5,538,889 | Peft-3 | GFP | oxTi880 | EG8837 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi880 IV | TAACTGGAGTGAATAAAGGAATCTCTCTTCTTC | In drp-1 | |
| LG4 | IV:11.69 | 14,278,434 | Peft-3 | GFP(NLS) | oxTi971 | EG8914 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi971 IV | NAAGTTTAATATAGAGTAGGAATATTGAATTGAA | In Y64G10A.6 | |
| LG4 | IV:12.29 | 14,665,718 | Peft-3 | tdTomato:H2B | oxTi391 | EG7933 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi391 IV | TATTGTTGAGTTCCAAAAGTTATTGAGACG | In anoh-2 | |
| LG4 | IV:14.22 | 15,587,320 | Peft-3 | GFP(NLS) | oxTi912 | EG8866 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi912 IV | TACTGATAAAGAGGAAGCTACTAATTCGTCCTTCAT | Intergenic (piRNA cluster) | |
| LG4 | IV:14.35 | 15,997,713 | Peft-3 | GFP | umnTi7 | cbr-unc-119(+) | unc-119(ed3) III; umnTi7 IV | Generated by the CGC | |||
| LG4 | IV:15.09 | 16,494,495 | Peft-3 | tdTomato:H2B | oxTi554 | EG7934 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi554 IV | TAGATAGATATAGATGAGATGAAGAGCGTT | In Y51H4A.2 | |
| LG4 | IV:15.17 | 16,635,091 | Peft-3 | GFP(NLS) | oxTi1000 | EG8939 | PuroR | oxTi1000 IV | TATGTGCATGTGCAAAATCAACAAAAACTCCAGTAAA | Intergenic (piRNA cluster) | Correction Feb 8, 2017: Genotype corrected to indicate insertion into N2 and NOT unc-119(ed3) |
| LG4 | IV:16.64 | 17,387,639 | Peft-3 | tdTomato:H2B | oxTi581 | EG7935 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi581 IV | TAAACAATTTATCTTTGAATTTATATCTAA | Intergenic | |
| LG4 | IV:2.45 | 5,876,776 | Peft-3 | GFP | oxTi208 | EG7916 | cbr-unc-119(+) and NeoR and Phsp:peel-1 | unc-119(ed3) III ; oxTi208 IV | TAGTTTTTGATTGATGCAATGTAACCGTGT | In srv-21 | |
| LG4 | IV:2.45 | 5,881,662 | Peft-3 | GFP(NLS) | oxTi994 | EG8933 | PuroR | oxTi994 IV | TATCTCTGAATAGTCATTAGTAGGCAATTCCATTACA | Intergenic (pseudogene) | Correction Feb 8, 2017: Genotype corrected to indicate insertion into N2 and NOT unc-119(ed3) |
| LG4 | IV:2.79 | 5,956,742 | Peft-3 | GFP(NLS) | oxTi938 | EG8889 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi938 IV | TATTAACAAGCTTTCACTGATTCCATATCCATCTGC | Intergenic (piRNA cluster) | |
| LG4 | IV:3.20 | 6,403,618 | Peft-3 | GFP(NLS) | oxTi1017 | EG8953 | NeoR | oxTi1017 IV | TAATTGCAAATCACCTGCTCAAATTCTTCAAATTT | Intergenic (piRNA cluster) | Correction Feb 8, 2017: Genotype corrected to indicate insertion into N2 and NOT unc-119(ed3) |
| LG4 | IV:3.22 | 6,707,019 | Peft-3 | tdTomato:H2B | oxTi616 | EG7917 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi616 IV | TATATTGTAGAGAAAACTTTATTAAAACTG | Intergenic | |
| LG4 | IV:3.22 | 6,673,721 | Peft-3 | GFP(NLS) | oxTi949 | EG8898 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi949 IV | TAGAACACGATGGCTCGTTTTTCAAGCTATTGAA | Intergenic | |
| LG4 | IV:3.23 | 6,785,258 | Peft-3 | GFP(NLS) | oxTi988 | EG8927 | HygroR | oxTi988 IV | TACTCTTTTTACAGCTCTCTCTAACTA | Intergenic | Correction Feb 8, 2017: Genotype corrected to indicate insertion into N2 and NOT unc-119(ed3) |
| LG4 | IV:3.24 | 6,810,857 | Peft-3 | tdTomato:H2B | oxTi727 | EG7918 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi727 IV | TAAACTATCAAAAAAAAACTAAATTAAAAA | Intergenic | |
| LG4 | IV:3.29 | 7,175,624 | Peft-3 | tdTomato:H2B | oxTi584 | EG7919 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi584 IV | TAAATTACAGATGACGTCTCTGTCTGAGAT | Intergenic | |
| LG4 | IV:3.71 | 8,352,355 | Peft-3 | tdTomato:H2B | oxTi551 | EG7920 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi551 IV | TACAGAATGAAGTGGAAAAAATTCGAACCA | Intergenic | |
| LG4 | IV:3.75 | 8,385,741 | Peft-3 | GFP(NLS) | oxTi1008 | EG8946 | NeoR | oxTi1008 IV | TACTTTATGGTTACGACTNAAAACTTTAACATATTT | Intergenic | Correction Feb 8, 2017: Genotype corrected to indicate insertion into N2 and NOT unc-119(ed3) |
| LG4 | IV:3.84 | 8,476,790 | Peft-3 | mCherry | oxTi414 | EG7921 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi414 IV | TAAAATAGTCAGACAACTGCTCTCATGCTA | Intergenic | |
| LG4 | IV:3.90 | 8,576,890 | Pvha-6 | GFP(NLS) | oxTi884 | EG8839 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi884 IV | TATATCGTATCGTAAACAGCTATATCGTACCGTTTA | Intergenic | |
| LG4 | IV:4.02 | 9,056,986 | Peft-3 | GFP(NLS) | oxTi909 | EG8863 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi909 IV | TACGTAAGTGGCACATTTACTTCGACGTCTAATTT | Intergenic | |
| LG4 | IV:4.05 | 9,112,907 | Peft-3 | GFP(NLS) | oxTi1023 | EG8959 | NeoR | oxTi1023 IV | TATGGTAGCTCACCACTTACAGTGTCAGTGCCAAT | Intergenic | Correction Feb 8, 2017: Genotype corrected to indicate insertion into N2 and NOT unc-119(ed3) |
| LG4 | IV:4.29 | 9,507,419 | Peft-3 | tdTomato:H2B | oxTi537 | EG7922 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi537 IV | TATGTCCTGGAGTCTCTCTCACTTCTAATT | In ethe-1 | |
| LG4 | IV:4.32 | 9,571,070 | Peft-3 | tdTomato:H2B | oxTi552 | EG7923 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi552 IV | TATAACGTGCTCAAAAGCTGGAGTATCGGC | In npr-2 | |
| LG4 | IV:4.34 | 9,617,514 | Peft-3 | tdTomato:H2B | oxTi599 | EG7924 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi599 IV | TATTAGAGTCGGCGGTAGGTATCCCTCTCT | In elt-1 | |
| LG4 | IV:4.38 | 9,711,782 | Peft-3 | tdTomato:H2B | oxTi611 | EG7925 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi611 IV | TATTTGGCAGAAACAGTCTATCTAGTTGGA | In nhr-38 | |
| LG4 | IV:4.40 | 9,744,069 | Peft-3 | mCherry | oxTi418 | EG7926 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi418 IV | TATAGTGGAGTCAATACACATTCACCAGAT | In C28D4.4, 5 or 7 | Non unique. 9,744,069 or 9,745,893 or 9,747,508 on ChrIV. |
| LG4 | IV:4.47 | 10,023,104 | Peft-3 | tdTomato:H2B | oxTi404 | EG7927 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi404 IV | TAGACATGTGGTAAATACAATTATGAACTA | Intergenic | |
| LG4 | IV:4.53 | 10,137,408 | Peft-3 | GFP | oxTi876 | EG8833 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi876 IV | TAAGACAGACATAAAAGCGGTCTTGGTTCTATTT | Intergenic | |
| LG4 | IV:4.62 | 10,509,881 | Peft-3 | GFP(NLS) | oxTi1014 | EG8950 | NeoR | oxTi1014 IV | TATTAATAGGTCTTGAAATGGTATTCTATATGAAA | Intergenic | Correction Feb 8, 2017: Genotype corrected to indicate insertion into N2 and NOT unc-119(ed3) |
| LG4 | IV:4.64 | 10,627,258 | Peft-3 | tdTomato:H2B | oxTi407 | EG7928 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi407 IV ; him-5(e1490) V | TATTTGAATGTTTTAAATATAAATTTTATT | In F09H10.5 | |
| LG4 | IV:4.92 | 11,188,673 | Peft-3 | GFP(NLS) | oxTi970 | EG8913 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi970 IV | ATTTGGGTTTAATCCCGGACTTCCTAATGAGAC | Intergenic | |
| LG4 | IV:5.02 | 11,589,911 | Peft-3 | tdTomato:H2B | oxTi602 | EG7929 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi602 IV | TATTCCAAAAAAACCAACCACAACTTCACT | In F40F11.4 | |
| LG4 | IV:5.45 | 11,958,382 | Peft-3 | tdTomato:H2B | oxTi538 | EG7930 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi538 IV | TAAATCATTACAGTTTTGCAAATAAACGGA | In tgt-1 | |
| LG4 | IV:5.46 | 11,983,669 | Peft-3 | tdTomato:H2B | oxTi577 | EG7931 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi577 IV | TACATTGAACTGGAAGCCTCCAACTGATAA | In unc-22 | |
| LG4 | IV:6.44 | 12,874,977 | Peft-3 | GFP | oxTi873 | EG8830 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi873 IV | TANNTTGAGTATAACTATACATAAAATCTTCTAA | In ZK896.1 | |
| LG4 | IV:6.87 | 13,023,274 | Peft-3 | GFP(NLS) | oxTi996 | EG8935 | PuroR | oxTi996 IV | TATATCATTTCAATTATTCACTCGTTTCTTCCCA | In mbk-2 | Correction Feb 8, 2017: Genotype corrected to indicate insertion into N2 and NOT unc-119(ed3) |
| LG4 | IV:8.43 | 13,177,428 | Peft-3 | tdTomato:H2B | oxTi592 | EG7932 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi592 IV | TACTTATTGAAAAGAACAATATCTGGAAGC | In lev-1 | |
| LG4 | IV:8.48 | 13,215,045 | Peft-3 | GFP | umnTi5 | cbr-unc-119(+) | unc-119(ed3) III; umnTi5 IV | Generated by the CGC | |||
| LG4 | IV:8.85 | 13,383,508 | Peft-3 | GFP(NLS) | oxTi921 | EG8873 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi921 IV | TACTTGCAAGCAATCATTTGGAAGTCTTCAGAGAT | In rab-19 | |
| LG5 | V:-0.18 | 6,417,733 | Peft-3 | tdTomato:H2B | oxTi703 | EG7943 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi703 V | TAACTCCCTAGTTAATCCATTTTCGCTTTT | In T05H4.10 | |
| LG5 | V:-1.51 | 5,573,238 | Peft-3 | tdTomato:H2B | oxTi392 | EG7565 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi392 V | TATGTACTTGGTTTTATCGTGAGAACGTGC | Intergenic | |
| LG5 | V:-12.80 | 2,968,566 | Peft-3 | tdTomato:H2B | oxTi654 | EG7939 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi654 V | TATTTCCTTAGTTACCTTTCATTATAAGTT | Intergenic | |
| LG5 | V:-12.80 | 2,969,544 | Peft-3 | tdTomato:H2B | oxTi708 | EG7940 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi708 V | TATTTAAATTTTTAAAAAATCAATCTAAAC | In pseudogene C44C3.4 | |
| LG5 | V:-13.00 | 2,820,016 | Peft-3 | GFP | oxTi874 | EG8831 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi874 V | TATCTGCGAAATAGCGTTTTGGGTTTTATCGAAA | In srh-36 | |
| LG5 | V:-18.05 | 1,614,903 | Peft-3 | GFP(NLS) | oxTi980 | EG8921 | HygroR | oxTi980 V | TAATGAGACAAGCTCTCACGCCTGGTGCGTAA | Intergenic (srbc-41 promoter) | Correction Feb 8, 2017: Genotype corrected to indicate insertion into N2 and NOT unc-119(ed3) |
| LG5 | V:-19.78 | 1,312,715 | Peft-3 | tdTomato:H2B | oxTi640 | EG7938 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi640 V | TAATTGTATTTTCGAATCTGTTTTTCTCGC | Intergenic | |
| LG5 | V:-19.85 | 1,247,232 | Peft-3 | GFP(NLS) | oxTi973 | EG8915 | HygroR | oxTi973 V | TAGAAGAACTTGAGAAAATGAGAAAAATATATA | Intergenic | Correction Feb 8, 2017: Genotype corrected to indicate insertion into N2 and NOT unc-119(ed3) |
| LG5 | V:-19.95 | 875,029 | Peft-3 | GFP(NLS) | oxTi1006 | EG8944 | PuroR | oxTi1006 V | TATACCCTTANAGAGCAGAATTTCCACTAAAA | Intergenic (str-53 pseudogen) | Correction Feb 8, 2017: Genotype corrected to indicate insertion into N2 and NOT unc-119(ed3) |
| LG5 | V:-2.39 | 5,192,031 | Peft-3 | GFP | oxTi211 | EG7566 | cbr-unc-119(+) and NeoR and Phsp:peel-1 | unc-119(ed3) III ; oxTi211 V | TATTTTAGGATGTTGTGATTGTTCGAGTCA | In C03A7.13 | |
| LG5 | V:-2.52 | 5,099,279 | Peft-3 | GFP(NLS) | oxTi965 | EG8908 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi965 V | NAATATTCGTGAAACTGCAATTTTCAACTG | Intergenic | |
| LG5 | V:-2.60 | 5,095,760 | Peft-3 | GFP(NLS) | oxTi1020 | EG8956 | NeoR | oxTi1020 V | TAATTGGTAATTTTTAATCTGAAATATAAA | In C04F5.2 | Correction Feb 8, 2017: Genotype corrected to indicate insertion into N2 and NOT unc-119(ed3) |
| LG5 | V:-20.00 | 307,492 | Peft-3 | tdTomato:H2B | oxTi664 | EG7936 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi664 V | TAAAACAGATTTTAGAACAATGGAAAATTT | In phf-10 | |
| LG5 | V:-20.00 | 380,392 | Peft-3 | GFP(NLS) | oxTi947 | EG8897 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi947 V | TNTGTGTAAGTAACTAATTTGGGTCATTTGAG | Intergenic | |
| LG5 | V:-4.37 | 4,748,464 | Peft-3 | tdTomato:H2B | oxTi393 | EG7942 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi393 V | TAAAATGTATACATTCACTGGTTCAGTTTG | Intergenic | |
| LG5 | V:-6.19 | 4,266,819 | Peft-3 | tdTomato:H2B | oxTi396 | EG7941 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi396 V | TAGTTGTAAGTGTTTTGCGTATAAAAAATT | In srt-36 or srt-37. | Non-unique. 4,266,819 or 4,280,292 on Chr. V. |
| LG5 | V:0.29 | 6,809,691 | Peft-3 | tdTomato:H2B | oxTi553 | EG7944 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi553 V | TACTTTTTCTTTTAATCTACTAAACATATA | Intergenic | |
| LG5 | V:0.65 | 7,315,737 | Peft-3 | tdTomato:H2B | oxTi543 | EG7945 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi543 V | TATTCGCATTTGTTGCGATGTATGAGGATG | In F07G11.9 | |
| LG5 | V:1.44 | 8,545,431 | Pmyo-3 | GFP(NLS) | oxTi889 | EG8844 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi889 V | TACATCAATTCCAGCTGCTACTTCAGTGCG | In F25G6.7 | |
| LG5 | V:1.58 | 8,748,594 | Peft-3 | GFP | oxTi878 | EG8835 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi878 V | TATTAAACGTCACAGTTTGCACTCCGAATACGT | In B0507.6 | |
| LG5 | V:1.91 | 9,046,227 | Peft-3 | tdTomato:H2B | oxTi580 | EG7946 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi580 V | TATTCTGCCACAAAGATTTGAAAATGCGGG | In nhr-30 | |
| LG5 | V:11.54 | 17,005,831 | Peft-3 | GFP(NLS) | oxTi894 | EG8849 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi894 V | TATAGTTGGAATGTCGATTCTCGGATTCTTTGAG | In srh-131 | |
| LG5 | V:11.98 | 17,031,532 | Peft-3 | GFP(NLS) | oxTi907 | EG8861 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi907 V | TAAACTTAATTCAGGTTCCAAAATTTCTCGGATC | in srz-92 (pseudogen) | |
| LG5 | V:12.85 | 17,303,245 | Peft-3 | GFP(NLS) | oxTi908 | EG8862 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi908 V | TACAACCTGAAATTGTATAAAACTTATACGTATC | In sri-70 | |
| LG5 | V:12.90 | 17,431,647 | Peft-3 | GFP(NLS) | oxTi963 | EG8907 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi963 V | NATGCAGACGTCCGAAATGAACGCGACTAAGA | In srbc-27 | |
| LG5 | V:13.04 | 17,706,897 | Peft-3 | GFP(NLS) | oxTi961 | EG8905 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi961 V | TATGTATTGTGAAATATAAAGCACTTTATTTCAT | Intergenic | |
| LG5 | V:18.45 | 18,862,976 | Peft-3 | GFP(NLS) | oxTi966 | EG8909 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi966 V | TAGATATTAACATCAAGTTTCTAACGGCTAAC | In emb-4 | |
| LG5 | V:19.80 | 19,092,734 | Peft-3 | GFP(NLS) | oxTi901 | EG8855 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi901 V | TAATAAAAGTTTATATTGATTGTTCAGAAATTTTT | Integenic | |
| LG5 | V:2.02 | 9,435,016 | Peft-3 | tdTomato:H2B | oxTi606 | EG7947 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi606 V | TAAATGGTCGTTTTCATTTAATAATAAAAT | Intergenic. | Non-Unique. 9,428,595 or 9,428,809 or 9,429,451 or 9,429,879 or 9,430,093 or 9,431,591 or 9,435,016 on Chr. V. |
| LG5 | V:2.02 | 9,446,802 | Peft-3 | GFP | oxTi879 | EG8836 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi879 V | TATTTGTCTCCAGTTAATGCATTAACTATTTCATT | In H24G06.1 | |
| LG5 | V:2.06 | 9,654,025 | Peft-3 | GFP(NLS) | oxTi997 | EG8936 | PuroR | oxTi997 V | TATATGTATATGGTAGGTCGTGAAAATCTAAATC | Intergenic | Correction Feb 8, 2017: Genotype corrected to indicate insertion into N2 and NOT unc-119(ed3) |
| LG5 | V:2.90 | 11,033,342 | Peft-3 | mCherry | oxTi417 | EG7309 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi417 V | TAAATGACACATACCAACCTTAACGGTTGC | Intergenic | |
| LG5 | V:2.92 | 11,050,601 | Peft-3 | tdTomato:H2B | oxTi620 | EG7948 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi620 V | TATACCTGTTGTTTATCGGAATCATAATTT | In pseudogene C45B11.7 | |
| LG5 | V:20.19 | 19,102,373 | Peft-3 | GFP | oxTi875 | EG8832 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi875 V | TAAAAATTGGGAAAAAATGCAAATTTTCTG | pro-3 3'UTR | |
| LG5 | V:20.20 | 19,130,943 | Peft-3 | GFP(NLS) | oxTi987 | EG8926 | HygroR | oxTi987 V | TAGAAAACCAAACTTCGAAAAAAAAATCAAT | In dyf-17 | Correction Feb 8, 2017: Genotype corrected to indicate insertion into N2 and NOT unc-119(ed3) |
| LG5 | V:24.52 | 20,056,389 | Peft-3 | GFP(NLS) | oxTi906 | EG8860 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi906 V | TAAATTCATGCAAAATTTTGCGTAGATTTC | In ttx-1 | |
| LG5 | V:24.99 | 20,445,995 | Peft-3 | tdTomato:H2B | oxTi633 | EG7968 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi633 V | TATAAATGTTTCCAGGAACTTCCGCCAATC | In pseudogene F48F5.2 | |
| LG5 | V:3.19 | 11,535,674 | Peft-3 | tdTomato:H2B | oxTi635 | EG7949 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi635 V | TAGAAGCAAATGTTGCCATTTCGATTGCTG | In ccch-1 | |
| LG5 | V:3.38 | 11,775,848 | Peft-3 | tdTomato:H2B | oxTi711 | EG7950 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi711 V | TAGTTGTTCTTTAATCGTCCATCGACTAAA | In bus-18 | |
| LG5 | V:3.38 | 11,808,281 | Peft-3 | GFP(NLS) | oxTi933 | EG8885 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi933 V | TAAATGTGTATTATTGGGAATAGGCCCATTC | In D2023.3 | |
| LG5 | V:3.42 | 11,832,160 | Peft-3 | tdTomato:H2B | oxTi630 | EG7951 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi630 V | TAGTTTGAACGGATTTTTTACCTTATTCTG | In nphp-4 | |
| LG5 | V:3.88 | 12,236,230 | Pmyo-3 | GFP(NLS) | oxTi888 | EG8843 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi888 V | TAGTTCATCACATAATCCTGAAAACACTGA | Intergenic | |
| LG5 | V:3.96 | 12,262,015 | Peft-3 | GFP | oxTi207 | EG7952 | cbr-unc-119(+) and NeoR and Phsp:peel-1 | unc-119(ed3) III ; oxTi207 V | TATGTTCCTCGTGCGATTTTGGTAGATCTT | In tbb-6 | |
| LG5 | V:4.26 | 12,456,908 | Peft-3 | tdTomato:H2B | oxTi542 | EG7953 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi542 V | TAATGTAATACTCACCGCTGCTTCATCAAA | In egl-10 | |
| LG5 | V:4.72 | 12,879,465 | Peft-3 | tdTomato:H2B | oxTi575 | EG7954 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi575 V | TATAAACAAACGAAAAACAACTCACCAATA | In bus-19 | |
| LG5 | V:4.95 | 13,139,970 | Peft-3 | GFP(NLS) | oxTi911 | EG8865 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi911 V | TAATAATCAATTTCGCTCTATCATCCAAATC | In dhc-3 | |
| LG5 | V:5.35 | 13,474,918 | Peft-3 | tdTomato:H2B | oxTi725 | EG7955 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi725 V | TACTTATTATCTTCAGCCCTGGCATTGAAC | In T10G3.8 | |
| LG5 | V:5.53 | 13,618,427 | Peft-3 | GFP(NLS) | oxTi1007 | EG8945 | NeoR | oxTi1007 V | TATGTTCTAGGTTCCTAGGATC | In srd-11 | Correction Feb 8, 2017: Genotype corrected to indicate insertion into N2 and NOT unc-119(ed3) |
| LG5 | V:5.54 | 13,628,082 | Peft-3 | tdTomato:H2B | oxTi547 | EG7956 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi547 V | TAAGACATTGGAGTTCATCCGAACTAATTT | In F53F4.14 | |
| LG5 | V:5.59 | 13,720,514 | Peft-3 | GFP(NLS) | oxTi991 | EG8930 | HygroR | oxTi991 V | TAAATTATTCAAAAAAACATATTTAATTTTTC | Intergenic | Correction Feb 8, 2017: Genotype corrected to indicate insertion into N2 and NOT unc-119(ed3) |
| LG5 | V:5.99 | 14,015,391 | Peft-3 | tdTomato:H2B | oxTi389 | EG7957 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi389 V | TAAGTTTGAATTATTTTGAATGTTCTATTT | In end-3 | |
| LG5 | V:6.23 | 14,310,043 | Peft-3 | GFP(NLS) | oxTi998 | EG8937 | PuroR | oxTi998 V | TACAAAATAATGTAAACATACCAGTTGGGTATT | In F14D7.14 | Correction Feb 8, 2017: Genotype corrected to indicate insertion into N2 and NOT unc-119(ed3) |
| LG5 | V:6.31 | 14,432,584 | Peft-3 | tdTomato:H2B | oxTi710 | EG7958 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi710 V | TATACAGTTTCGCAATAACCTCCCCCGTGC | Intergenic | |
| LG5 | V:6.72 | 14,721,283 | Peft-3 | GFP(NLS) | oxTi999 | EG8938 | PuroR | oxTi999 V | TANATGGTGCCACATGAAGTTTGTAGGCAACT | Intergenic | Correction Feb 8, 2017: Genotype corrected to indicate insertion into N2 and NOT unc-119(ed3) |
| LG5 | V:6.85 | 14,778,098 | Peft-3 | GFP(NLS) | oxTi1021 | EG8957 | NeoR | oxTi1021 V | TAATCCNTGNAGACTCACCATTTTACATTCAA | Intergenic | Correction Feb 8, 2017: Genotype corrected to indicate insertion into N2 and NOT unc-119(ed3) |
| LG5 | V:8.58 | 15,663,652 | Peft-3 | GFP(NLS) | oxTi922 | EG8874 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi922 V | TATCTAACATTAAATAAATCAATTTTCATTCTA | Intergenic | |
| LG5 | V:8.97 | 15,838,575 | Peft-3 | tdTomato:H2B> | oxTi405 | EG7959 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi405 V him-5(e1490) V | TACTGTGGGATGGAGTAGCGTCAGTGGAGA | In pseudogene F54B8.13 | |
| LG5 | V:9.11 | 15,894,022 | Peft-3 | tdTomato:H2B | oxTi401 | EG7960 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi401 V | TATCCTGCATTTGAACCTTTAA | In T06C12.11 | |
| LGX | X:-0.43 | 8,160,889 | Peft-3 | GFP(NLS) | oxTi890 | EG8845 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi890 X | TAAGTGAGGAGTAGTATTTTTGCAAAATTATT | Intergenic | |
| LGX | X:-1.86 | 7,108,094 | Peft-3 | tdTomato:H2B | oxTi704 | EG7988 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi704 X | TAGTTATTACAAATTTAAAAAAGACTGCAA | In C36B7.2 | |
| LGX | X:-13.81 | 2,702,823 | Peft-3 | GFP(NLS) | oxTi936 | EG8888 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi936 X | TACTGTGTACAGTTCTTTTTTAATAACCACCT | In igcm-3 | |
| LGX | X:-14.14 | 2,629,820 | Peft-3 | tdTomato:H2B | oxTi675 | EG7978 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi675 X | TATACATAGTAAGAATGACGACAAAAACTG | In F54G2.1 | |
| LGX | X:-14.39 | 2,563,224 | Peft-3 | tdTomato:H2B | oxTi410 | EG7977 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi410 X | TAGGCTTAGGCTTTGGCTTTGGCGTAGGCT | In F52H2.6 | |
| LGX | X:-16.90 | 2,188,310 | Peft-3 | tdTomato:H2B | oxTi702 | EG7976 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi702 X | TACATATAAATATATTTATATGTCAATGGG | Intergenic | |
| LGX | X:-17.06 | 2,080,345 | Peft-3 | tdTomato:H2B | oxTi651 | EG7975 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi651 X | TAGACATGGTTGCAGACGAATCTTCTGACG | In F49H12.4 | |
| LGX | X:-18.91 | 949,571 | Peft-3 | tdTomato:H2B | oxTi673 | EG7972 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi673 X | TATGTCACAATTGTGTCGTCTCTCATTCTC | Intergenic | |
| LGX | X:-19.89 | 436,809 | Peft-3 | tdTomato:H2B | oxTi406 | EG7971 | cbr-unc-119(+) | unc-119(ed3) III ; him-5(e1490) V ; oxTi406 X | TACACTGTTATGGACAGATATGAAAGCAGG | Intergenic | |
| LGX | X:-2.45 | 6,760,762 | Peft-3 | GFP(NLS) | oxTi953 | EG8900 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi953 X | TAAGTAGGGTGTAATAATTTTCCACTGGGCCGT | In abts-4 | |
| LGX | X:-2.56 | 6,685,324 | Peft-3 | mCherry | oxTi416 | EG7987 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi416 X | TAAACAATGTGAAATTCAGACTAGATCCAT | In F38B6.6 | |
| LGX | X:-20.11 | 366,632 | Peft-3 | tdTomato:H2B | oxTi597 | EG7970 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi597 X | TAAAGGGATTTTAACAACTTTTTATAAATG | Intergenic | |
| LGX | X:-20.17 | 333,709 | Peft-3 | tdTomato:H2B | oxTi621 | EG7969 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi621 X | TATGTAAGTTGAAAGATTTTTAAAAAAAGT | Intergenic | |
| LGX | X:-20.67 | 170,242 | Peft-3 | GFP(NLS) | oxTi986 | EG8925 | HygroR | oxTi986 X | TAGGTATATAGTTATATAGGTCTATAGGTTAT | In T08D2.2 | Correction Feb 8, 2017: Genotype corrected to indicate insertion into N2 and NOT unc-119(ed3) |
| LGX | X:-3.51 | 6,180,402 | Peft-3 | mCherry | oxTi421 | EG7986 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi421 X | TAAAGTTTCTGTGGATATGGAAAAAAATCA | Intergenic | |
| LGX | X:-3.89 | 6,010,872 | Peft-3 | GFP(NLS) | oxTi968 | EG8911 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi968 X | TAGAGTGGTCAGTGCTGTTGACCGTTTCGA | In C09B8.3 | |
| LGX | X:-4.88 | 5,545,540 | Peft-3 | tdTomato:H2B | oxTi566 | EG7985 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi566 X | TATAGGGAGCACAGATTGAAAACTCTCAAA | In C54G7.1 | |
| LGX | X:-7.13 | 4,628,593 | Peft-3 | GFP(NLS) | oxTi954 | EG8901 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi954 X | TATAGGTCCCTAGAGGTTTAATGTTTCGTAGA | Intergenic | |
| LGX | X:-7.36 | 4,522,755 | Peft-3 | GFP(NLS) | oxTi1019 | EG8955 | NeoR | oxTi1019 X | TATNTNTTTTTTTTCATATTGAAAAAGAAGT | Intergenic | Correction Feb 8, 2017: Genotype corrected to indicate insertion into N2 and NOT unc-119(ed3) |
| LGX | X:-7.77 | 4,348,071 | Peft-3 | tdTomato:H2B + gaIs283 | oxTi334 | EG7215 | cbr-unc-119(+) | oxTi334 X ; gaIs283 | TAAAAAGGGGTATTTTGGCACTTTCAAAAT | Intergenic | |
| LGX | X:-7.88 | 4,245,209 | Peft-3 | tdTomato:H2B + gaIs283 | oxTi335 | EG7216 | cbr-unc-119(+) | oxTi335 X ; gaIs283 | TATGTGTCTAAAATTAGTTAGATAGTATCA | In ncRNA F48D6.5 | |
| LGX | X:-8.62 | 4,031,634 | Peft-3 | tdTomato:H2B | oxTi728 | EG7982 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi728 X | TAGCTACTTACGATACGATATAGCTACTTA | Intergenic | Non unique. 4,031,634 or 4,037,579 or 4,048,874 Chr. X. |
| LGX | X:-9.13 | 3,913,671 | Pvha-6 | GFP(NLS) | oxTi885 | EG8840 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi885 X | TACATATGATTTTTCCTATCTAGCCGCTTTGT | Intergenic | |
| LGX | X:-9.30 | 3,841,631 | Peft-3 | tdTomato:H2B | oxTi639 | EG7981 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi639 X | TAATGGTGAAAAATAGCGAGTGTGAAAACG | In ttr-48 | |
| LGX | X:-9.39 | 3,796,520 | Peft-3 | GFP(NLS) | oxTi900 | EG8854 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi900 X | TAATTAACATCAAGGTATTAGTATTTATTGTGAAT | Intergenic | |
| LGX | X:-9.44 | 3,769,088 | Peft-3 | tdTomato:H2B | oxTi557 | EG7980 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi557 X | TATATTAGATTTTTTTCCCATATCTCTTGG | Intergenic | |
| LGX | X:0.19 | 8,670,153 | Peft-3 | tdTomato:H2B | oxTi668 | EG7989 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi668 X | TAAAGGCGAGCATTCGTCAACGTCGGCCTC | In rig-1 | |
| LGX | X:0.57 | 9,028,966 | Peft-3 | GFP(NLS) | oxTi893 | EG8848 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi893 X | TAGTACAGTTGATGGTTGTGAGAAATTTGNTT | Intergenic | |
| LGX | X:1.32 | 9,504,204 | Peft-3 | tdTomato:H2B + gaIs283 | oxTi333 | EG7214 | cbr-unc-119(+) | gaIs283 ; oxTi333 X | TATAGCCTTCATTCTTTCCTTCTTTCTATC | In gap-2 | |
| LGX | X:1.43 | 9,672,363 | Peft-3 | tdTomato:H2B | oxTi400 | EG7990 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi400 X | TACATAGTACATCAACAATCTGAAACCATT | Intergenic | |
| LGX | X:1.46 | 9,728,302 | Peft-3 | tdTomato:H2B | oxTi594 | EG7991 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi594 X | TAAATCCTTAAAAACGTTTTTATGGTGCTA | In him-4 | |
| LGX | X:1.87 | 10,169,225 | Peft-3 | tdTomato:H2B | oxTi545 | EG7992 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi545 X | TAATAAATTTTATGACAAAATATTGAGCAA | In tiar-3 | |
| LGX | X:12.63 | 13,482,789 | Peft-3 | tdTomato:H2B | oxTi395 | EG7994 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi395 X | TATTTTGAATATCTTCTCAACATATTCGGG | In srd-51 | |
| LGX | X:12.63 | 13,479,833 | Peft-3 | GFP(NLS) | oxTi1015 | EG8951 | NeoR | oxTi1015 X | TAAGCGGATTCAAATTAGGAACTAGCGAGAGT | In srd-50 | Correction Feb 8, 2017: Genotype corrected to indicate insertion into N2 and NOT unc-119(ed3) |
| LGX | X:16.41 | 14,085,719 | Peft-3 | tdTomato:H2B | oxTi596 | EG7995 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi596 X | TAAAGAGAAACGGTTTTGTTGGGATGTGGA | In F16B12.1 | |
| LGX | X:16.67 | 14,212,066 | Peft-3 | tdTomato:H2B | oxTi403 | EG7996 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi403 X | TAGAATGGCGACAAAAGGCTCAATTTCACA | Intergenic | |
| LGX | X:2.22 | 10,481,189 | Peft-3 | tdTomato:H2B | oxTi308 | EG7826 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi308 X | TATTATCCATTCTATAATCTATTTTCTAGT | Intergenic | |
| LGX | X:2.86 | 11,048,647 | Peft-3 | tdTomato:H2B | oxTi412 | EG7993 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi412 X | TATATCATCCATCCACTCACCCTCTTTGCA | In clc-1 | |
| LGX | X:22.87 | 15,582,062 | Peft-3 | tdTomato:H2B | oxTi709 | EG7998 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi709 X | TACATAAATACGTTAAAAAAAGTTACCCTG | In K09A9.6 | |
| LGX | X:23.69 | 16,055,776 | Peft-3 | tdTomato:H2B | oxTi649 | EG7999 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi649 X | TATTATGATATTCGGTCATGTTGGGACCAT | In T27A8.7 | |
| LGX | X:23.77 | 16,320,254 | Peft-3 | tdTomato:H2B | oxTi659 | EG8000 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi659 X | TAAATCAGGTTCAATGTGCACACCGCGACT | Intergenic | |
| LGX | X:23.96 | 16,427,094 | Peft-3 | tdTomato:H2B | oxTi701 | EG8001 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi701 X | TAGAGTTGTTTTCAAAAGTGCTCAAAAGTG | Intergenic | |
| LGX | X:24.08 | 17,154,612 | Peft-3 | GFP(NLS) | oxTi905 | EG8859 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi905 X | TAGGGTCAAAAATATCTTTAATAGTCAAACTGG | Intergenic | |
| LGX | X:24.09 | 17,288,880 | Peft-3 | GFP(NLS) | oxTi932 | EG8884 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi932 X | NAACCTTAAACCAATATCACTTCAAAAGAGGA | Intergenic | |
| LGX | X:24.09 | 17,233,242 | Peft-3 | GFP(NLS) | oxTi1003 | EG8942 | PuroR | oxTi1003 X | TATTTCGTACTTATTCATTTTACTTATACAAAAAA | In dhs-30 and T24G12.3 | Correction Feb 8, 2017: Genotype corrected to indicate insertion into N2 and NOT unc-119(ed3) |
| LGX | X:24.10 | 17,432,529 | Peft-3 | tdTomato:H2B | oxTi707 | EG8003 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi707 X | TATTACATCCCCTTATTTTGATGAATGTGT | In pbo-6 | |
| LGX | X:24.44 | 17,596,837 | Peft-3 | GFP(NLS) | oxTi898 | EG8852 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi898 X | TATTTCGAATTTTTCTCGAAAAACGGTTTTAAAA | Intergenic | |
| LGX | X:24.62 | 17,692,310 | Peft-3 | GFP(NLS) | oxTi910 | EG8864 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi910 X | TAGTTCGAAGCACCCTCCTTCCCAAAATCGTAG | Intergenic | |
| LGX | X:24.63 | 17,717,793 | Peft-3 | GFP(NLS) | oxTi899 | EG8853 | cbr-unc-119(+) | unc-119(ed3) III ; oxTi899 X | TACTGGAAAAATGTGGCAAAGCTTGGGTTTTTCG | In cTel55X.1 |
Note: Scroll right in table for genotype and flanking sequences.
Mapping strains
| Strain | Chr. | Genetic. pos. | Position (WS230) | Fluorophore | Allele | Insertion Marker |
|---|---|---|---|---|---|---|
| EG8040 | ||||||
| LG1 | I:4.7 | 10,166,145 | Peft-3:mCherry | oxTi302 | cb-unc-119(+) | |
| LG2 | II:-1.5 | 5,448,544 | Peft-3:GFP:H2B | oxTi75 | unc-18(+) | |
| LG3 | III:-1.4 | 6,076,837 | Peft-3:TdTomato:H2B | oxTi411 | cb-unc-119(+) | |
| him-8 | LG4 | |||||
| EG8041 | ||||||
| LG4 | IV:5.4 | 11,899,008 | Peft-3:GFP:H2B | oxTi76 | unc-18(+) | |
| LG5 | V:9.0 | 15,838,568 | Peft-3:TdTomato:H2B | oxTi405 | cb-unc-119(+) | |
| LG6 | X:-3.5 | 6,180,397 | Peft-3:mCherry | oxTi421 | cb-unc-119(+) | |
| him-5 | LG5 | |||||
Please note that strains containing the Peft-3:tdTomato:H2B are generally sick at high temperature (25°C).