Recent news
Wormbook chapter on C. elegans transgenic methods is out in Genetics.
Aug 23, 2019
My review / Wormbook chapter with Jeremy Nance (NYU) is out now in Genetics (paper here). Please email me if you don’t have access to Genetics and I will send you a pdf.
We try to cover the rapidly expanding set of tools that have become developed for C. elegans with a focus on recent developments. However, we’ve also tried to summarize some of the accumulated “wisdom” from many different labs to improve the chance of generating a functional transgene.
The different sections cover:
- C. elegans Genes and Transgene Structures
- Multicopy Transgenesis
- Integrating arrays
- Transposon-Mediated Genetic Engineering
- CRISPR/Cas9-Mediated Transgenesis
- Conditional Gene and Protein Inactivation
- Bipartite Systems for Temporal and Spatial Control of Expression
We hope the review will be a useful “entry point” for researchers that are new to C. elegans while also having information that for the seasoned C. elegans scientist.
Paper on non-Mendelian inheritance now published at Developmental Cell.
Feb 25, 2019
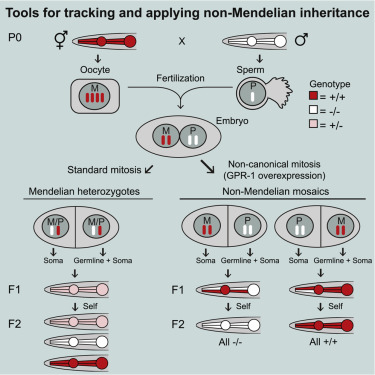
Karen’s paper on non-Mendelian inheritance is now online and can be found here. Please send me a request if you have trouble accessing the paper. In the paper, we generate a set of strains (available at cgc) that facilitate crosses where segregation follows a non-Mendelian pattern by overexpressing GPR-1, as shown by Besseling and Bringmann (2016). We demonstrate that the strains can be used to “transplant” mitochondria between strains and show lineage-specific requirements for vulval induction.
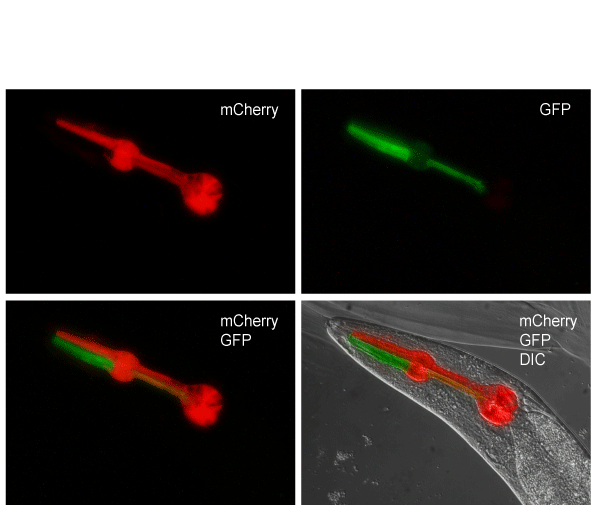
Other than making really cool pharyngeal expression patterns, the strains should be useful for determining maternal and paternal contributions to development and epigenetic, transgenerational inheritance.
The lab is growing!
Feb 25, 201

After a bit more than a year at KAUST, the lab now has two postdocs (Sonia el Mouridi and Amhed Velazquez), two Ph.D. students (Monika Priyardishini and Mohammed Aljohani), and one visiting student (Alhanouf).
New position as Assistant Professor at KAUST
Jan 2, 2018
I am excited to have recently started my own research group at KAUST (King Abdullah University of Science & Technology) in Saudi Arabia. KAUST is a very modern, international university with over 100 nationalities represented on campus, approximately 40% female students, and a focus on Engineering & Science. The university is near Jeddah on the coast of the Red Sea.
![]()
Although I realize Saudi Arabia is not the most obvious choice for establishing a laboratory, I was very attracted by the scientific community, the resources available for research, and about being part of “building” the academic environment at a young, ambitious university in a region, that is eager to educate their youth.
The laboratory is actively looking for postdocs, so if you are interested in developing large-scale genome engineering technologies with a focus on genome organization, then I encourage you to apply here.
The university has very attractive programs with all expenses paid to encourage undergraduate visitors. There is a poster presentation at the annual Winter Enrichment Program (this year, Human Machine Future). Also, there are month-long research projects with a stipend offered via the Visiting Student Research Program where we have a project listed. I strongly encourage an application; it is a great scientific and cultural experience, with some fun extracurricular activities thrown in together with the cohort of students (snorkeling and diving are spectacular here). So far, one female undergraduate student from a C. elegans lab in the US was “brave” enough to apply; she’ll come for WEP this January.
New Reagents Deposited with Addgene
Feb 8, 2017
We have deposited several plasmids with C. elegans optimized fluorophores with and without PATC. In our hands, including PATCs significantly diminishes epigenetic silencing in the germline but has no obvious positive or negative effect on somatic expression. The plasmids are described here and can be accessed at Addgene here.
Paper on Periodic An/Tn Clusters (PATCs) published
July 6, 2016
Our paper on epigenetic silencing of transgenes in the C. elegans germline just came out.
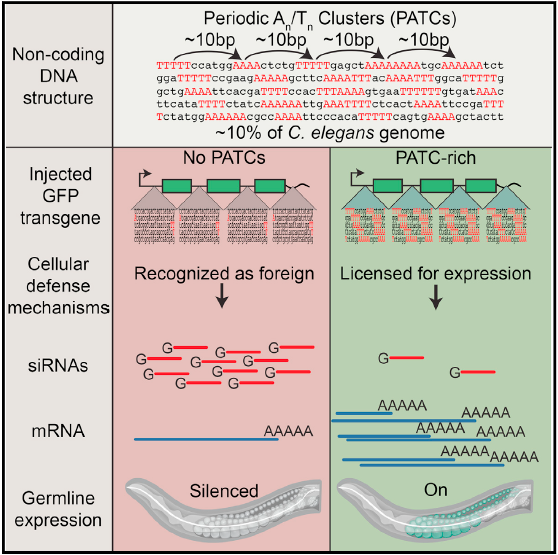
- Periodic, non-coding DNA can prevent transgenes from stochastic silencing in germline
- Non-coding content of genes is shaped by genomic context and heterochromatin domains
- Conditioning of active DNA may allow cells to distinguish foreign from host genes
The worm genome contains a prevalent (~10% of the genomic sequence) non-coding DNA feature called PATCs, which is highly enriched “around” genes expressed in the germline. PATCs are highly enriched in genes that reside in repressive chromatin environments, suggesting that this non-coding protects endogenous genes from epigenetic silencing. We propose that PATCs may also play a role in a basic cellular immune system to detect foreign DNA. In this model, germ cells use this abundant non-coding DNA as a signature to identify endogenous genes and to silence foreign DNA (transposons, retroviruses, transgenes) that lack PATCs by default. In line with this, we find that transgenes with PATCs are much less frequently silenced in the germline from permissive and repressive chromatin domains.
Fluorescent markers strains with GFP insertions.
In collaboration with Ann Rougvie at the CGC, we have generated a set of strains carrying GFP markers at defined genomic locations. The strains are available from the CGC and you can search for the strains based on chromosome, fluorescent marker, and the selection marker (see Fluorescent marker strains). The fluorescence is visible on a fluorescence dissection microscope and the strains can be used to build double mutants in crosses or for outcrossing mutations isolated in genetic screens. The strains complement the previously published set of strains with insertions of tdTomato and brings the total to 294 strains.
In addition, the CGC is generating fluorescently marked versions of some of the classic balancer strains by targeted integration using Cas9. We have generated a marked MT1000 strain (unc-5(e53)/nT1 IV; dpy-11(e224)/nT1 V) by inserting Peft-3:GFP into Chr. V at 2.8 MB. The strain can be used as a balancer even though the cytosolic GFP is somewhat dim. We have therefore switched to inserting a brighter Peft-3:tdTomato(NLS) marker.
Move to Andrew Fire’s lab at Stanford
Sept 16, 2014
I have just recently moved to Andrew Fire‘s lab at Stanford! In addition to developing techniques to engineer C. elegans, I will also work on understanding how cells in the germline identify foreign and endogenous genes as a postdoc in Andy’s lab (technically, a “visiting instructor”).
Thanks to Erik and everyone in the lab for a fantastic time at University of Utah!
Antibiotic selection protocol
July 2, 2014
I have added a page with the protocol we use in Erik’s lab for antibiotic selection.
Variable efficiency of peel-1 negative selection
June 17, 2014
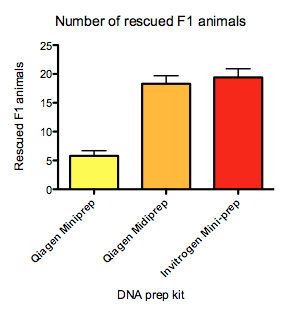
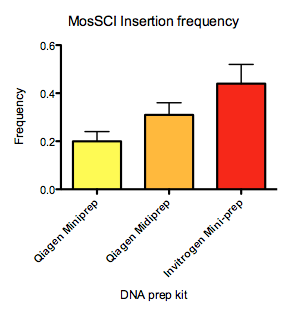
For no obvious reason, I have recently observed large variability in the efficiency in using the peel-1 negative selection to kill animals with extra-chromosomal arrays. When I initially characterized the negative selection (Frøkjær-Jensen et al., 2010) it was very efficient at killing essentially all animals with extra-chromosomal arrays. For a long time, I and others in Erik’s lab picked animals with mosSCI or miniMos insertions based on surviving the heat-shock and found essentially no false positives (by a secondary screen for mCherry co-injection markers). However for the past two summers, the negative selection has become very inefficient even when using the same injection mixes that previously worked. It is very strange but it coincides closely with the installation of a new evaporative cooling system in the building and the summer months. I have performed a number of experiments to enhance the efficiency of negative selection:
– Using the genomic peel-1 fragment instead of cDNA.
– Heat-shock in water bath instead of in an air incubator.
– Higher temperature heat-shock (1 hour at 37°C instead of 34°C).
– Doubling the concentration of the peel-1 plasmid (pMA122)
None of these changes improved the negative selection.
I apologize if others are observing similar variability. This was not something I observed when originally characterizing the negative selection. And if anyone has figured out how to reduce the variability (other than wait for fall and winter) then please send me an email. Or write up a paper on how “A sperm-delivered toxin can determine the seasons in the short-lived model organism C. elegans“…