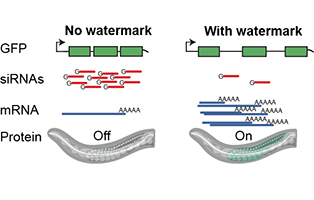
Genome organization and regulation of gene expression
We are interested in understanding the general design principles of how gene expression is regulated at the genomic level in multicellular organisms. This includes understanding how large-scale DNA characteristics such as patterns in non-coding sequence, gene structure, and large-scale chromosome organization influence gene expression. For example, given that a large fraction of genomes are derived from foreign DNA such as retrotransposons (~40% for the human genome), how do cells distinguish and differentially regulate the “self” and “foreign” genome? Similarly, if we consider the complete genomic sequence of an organism, what fraction of the DNA can be assigned defined function(s)?
To understand the grammar of how primary DNA sequence regulates gene expression, we use classical genetics, bioinformatics, and biological engineering in the nematode C. elegans, inspired by the nascent field of synthetic biology. We believe that quantitative, high-throughput approaches will drive insights into how gene expression is regulated in multi-cellular organisms, how developmental programs are organized, and how the primary genome sequence directs these programs. My lab will therefore have a dual focus: (1) to continuously improve and develop tools for true genome-scale engineering in C. elegans, and (2) to use these tools to identify and define the functional roles of regulatory DNA in the genome.
Developing methods for high efficiency genome editing in C. elegans
Development of high-throughput genome editing technologies. Quantifying functionality, as well as redundancy, of parts of the genome requires efficient genome editing. For example, what constitutes a minimal genome and number of genes that allows viability and normal development? Can we identify a set of enhancers and regulatory regions that allow expression in each individual cell at every developmental stage? Using transposons and Cas9/CRISPR, we will continue developing genome editing tools with the long-term goal of enabling genome-scale manipulation for arbitrarily complex re-organization and re-coding of the C. elegans genome to answer questions about genome structure and function.
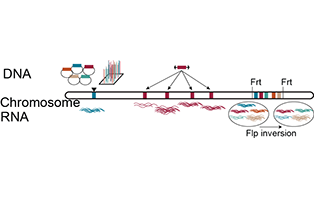
Sequence adaptations for expression
Which gene features ensure robust gene expression from different chromatin domains? Gene networks ensure invariant developmental outcomes despite noisy transcription and changes in chromatin environments, for example by cellular differentiation. We will use high-throughput insertion of natural and synthetic DNA fragments, as well as genome shuffling, to identify: (a) sequence features in enhancers and promoters that tune their expression to their chromatin environment, (b) genomic regions where gene silencing is developmentally regulated, and (c) gene structures (e.g. intron position and content) that regulate expression
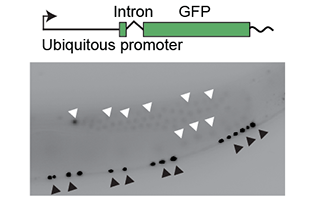
How do non-coding DNA structures allow cells to identify and silence foreign DNA?
Which gene features ensure robust gene expression from different chromatin domains? Gene networks ensure invariant developmental outcomes despite noisy transcription and changes in chromatin environments, for example by cellular differentiation. We will use high-throughput insertion of natural and synthetic DNA fragments, as well as genome shuffling, to identify: (a) sequence features in enhancers and promoters that tune their expression to their chromatin environment, (b) genomic regions where gene silencing is developmentally regulated, and (c) gene structures (e.g. intron position and content) that regulate expression